Histological image, because of its ex vivo and microscopic nature, usually provides structural or pathological ground-truth. Finding MR regions corresponding to histological ground-truth regions will help us better understand how a certain structure or pathology appears in MRI. This is often the building block for future labeling or diagnosing new subjects from their MR images.
Histology and MRI registration is in general a very challenging multi-modality registration task. Difficulties are:
Below we show an example of using DRAMMS to somehow alleviate some of those issues, mostly because of attribute matching design and mutual-saliency weighting components.
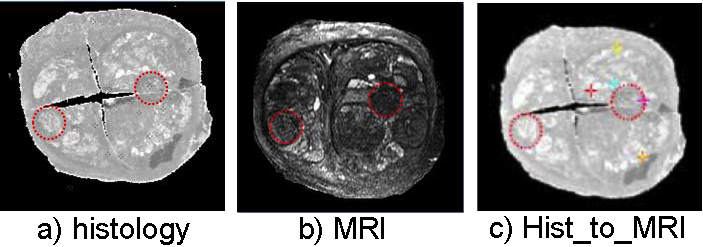
Registration of multi-modality images of a same prostate. Dashed circles outline the corresponding structures.
dramms -S src_histology.hdr -T trg_MRI.hdr
-O src2trg.nii.gz -D def_S2T.nii.gz
-g 0.25 -w 1 -c 2
Here ‘-w 1’ option tells dramms to use correlation coefficient (of the attribute vectors) as the similarity metric, instead of the default squared difference similarity metric. We recommend to use ‘-w 1’ option in multi-modality registration jobs.
Despite this successful example, registration of histological and MR images is very challenging task that even the affine registration may often fail. Because of the difficulty, DRAMMS may fail too.
One solution is to carefully re-do affine registration outside DRAMMS scope. After having obtained a reasonable affine result, one can input the affinely registered images (src2trg_affine.nii.gz and trg.nii.gz) into DRAMMS. Meanwhile bypassing the affine part within DRAMMS by the -a 0 option.
Registering this pair of 3D images (target image 256 x 256 x 64 voxels, 0.16 x 0.16 x 0.4 mm^3/voxel) takes 6.7 GB memory and finishes in 39 minutes in Linux OS (2.80GHz CPU).
If one can afford less memory, please use -u option to choose memory usage in different levels (the lowest being about 1/4 of maximum memory used). This may however slightly reduce registration accuracy.